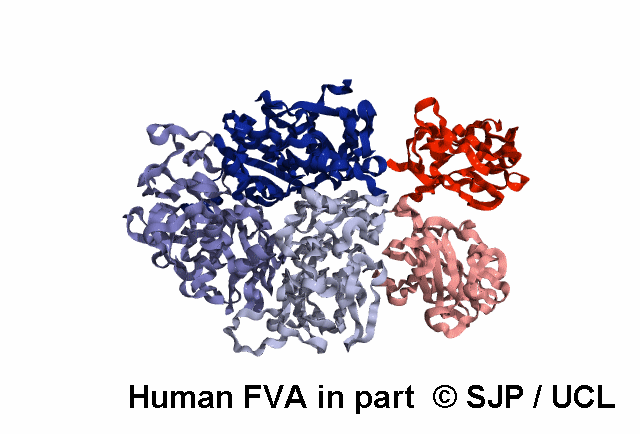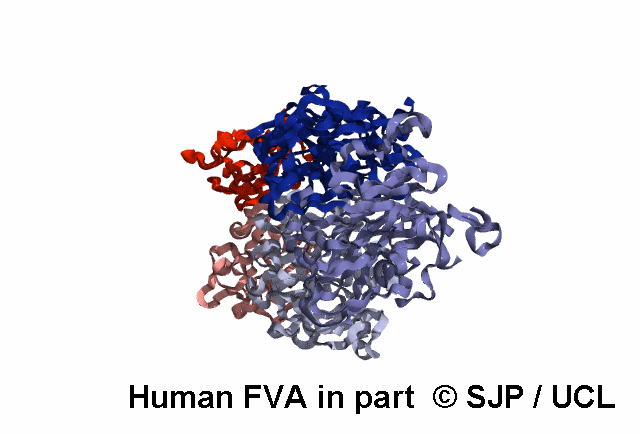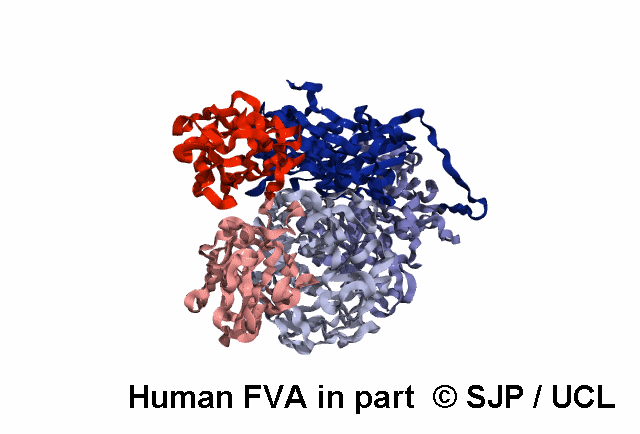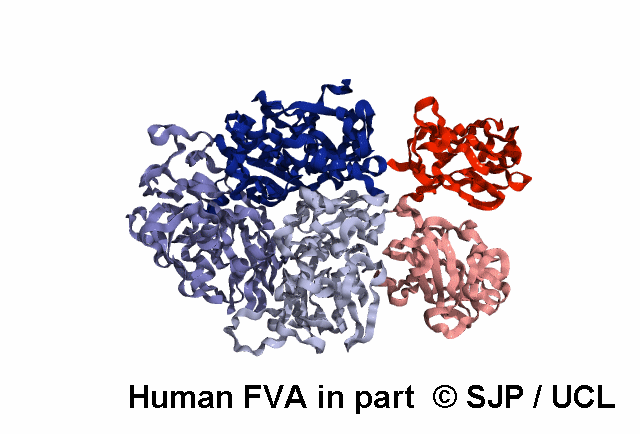
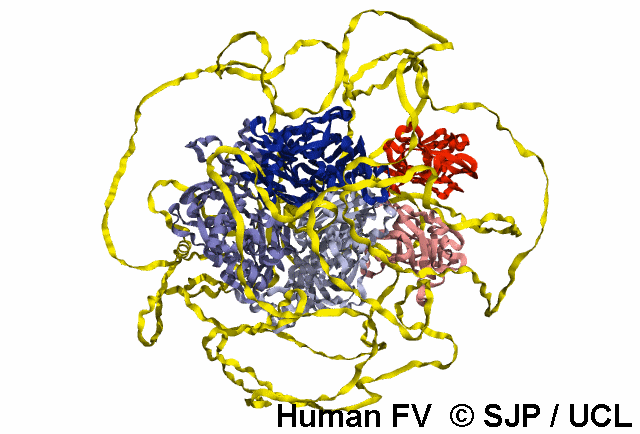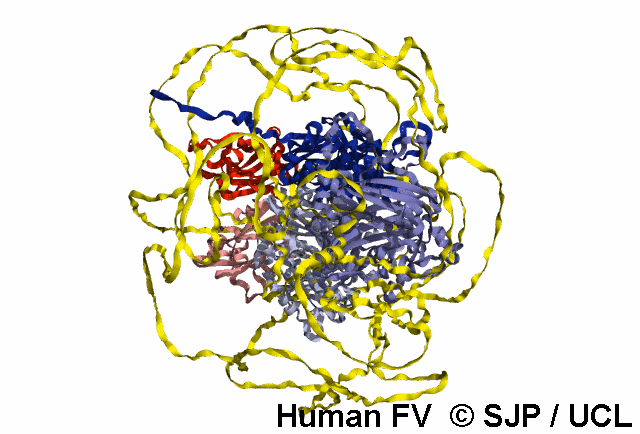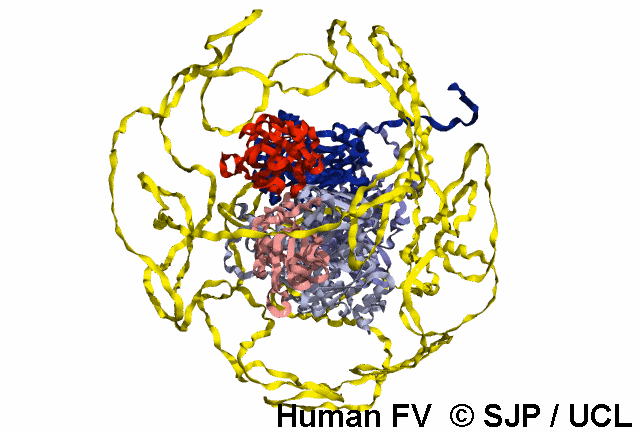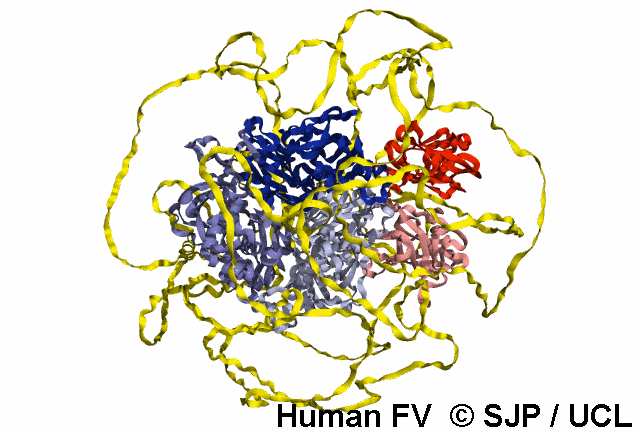
Structures available for Coagulation Factor V
All complete FV structural analysis presented within this database uses the previously published
7KVE
structure taken from the Protein Data Bank stitched with the AlphaFold FV prediction to fill in the missing B domain from 7KVE.
The complete FV structure consists of the A1, A2, B, A3, C1, and C2 domains.
FVa specific analysis uses the 7KXY PDB structure.
Experimental Structures available for Human Coagulation Factor V
PDB |
Method |
Description |
Resolution |
Chain |
Position |
PDBsum |
|---|---|---|---|---|---|---|
| 1CZS | X-RAY | CRYSTAL STRUCTURE OF THE C2 DOMAIN OF HUMAN COAGULATION FACTOR V: COMPLEX WITH PHENYLMERCURY | 1.9 | A | 2065-2224 | >> |
| 1CZT | X-RAY | CRYSTAL STRUCTURE OF THE C2 DOMAIN OF HUMAN COAGULATION FACTOR V | 1.8 | A | 2065-2224 | >> |
| 1CZV | X-RAY | CRYSTAL STRUCTURE OF THE C2 DOMAIN OF HUMAN COAGULATION FACTOR V: DIMERIC CRYSTAL FORM | 2.4 | A/B | 2065-2224 | >> |
| 3P6Z | X-RAY | Structural basis of thrombin mediated factor V activation: essential role of the hirudin-like sequence Glu666-Glu672 for processing at the heavy chain-B domain junction | 1.7 | C/F | 665-737 | >> |
| 3P70 | X-RAY | Structural basis of thrombin-mediated factor V activation: essential role of the hirudin-like sequence Glu666-Glu672 for processing at the heavy chain-B domain junction | 2.5 | I/J | 665-737 | >> |
| 3S9C | X-RAY | Russell's viper venom serine proteinase, RVV-V in complex with the fragment (residues 1533-1546) of human factor V | 1.8 | B | 1561-1574 | >> |
| 7KVE | ELECT | Cryo-EM structure of human Factor V at 3.3 Angstrom resolution | 3.3 | A | 29-2224 | >> |
| 7KVF | ELECT | Cryo-EM structure of human Factor V at 3.6 Angstrom resolution | 3.6 | A | 29-2224 | >> |
| 7KXY | ELECT | Cryo-EM structure of human Factor Va at 4.4 Angstrom resolution | 4.4 | A | 29-747 | >> |
| NA | PREDICT | Coagulation factor V AlphaFold structure prediction | NA | A | 1-2224 | >> |