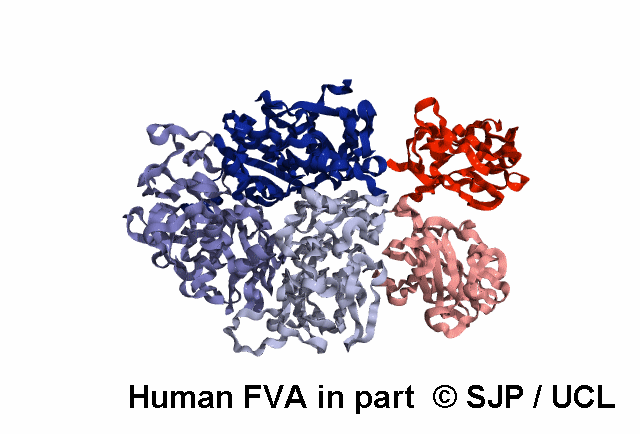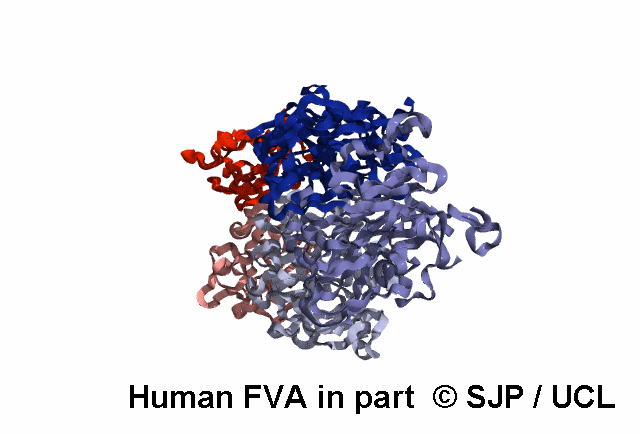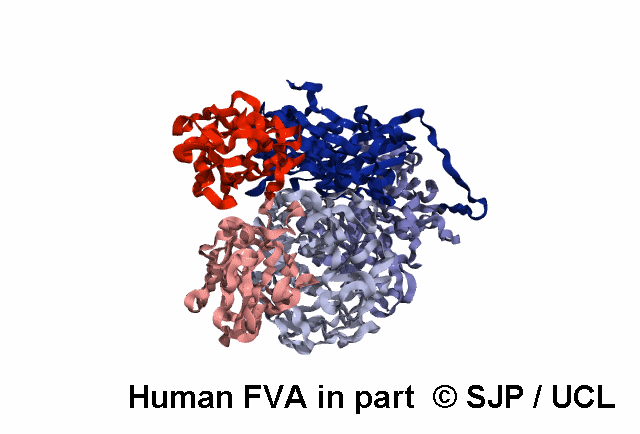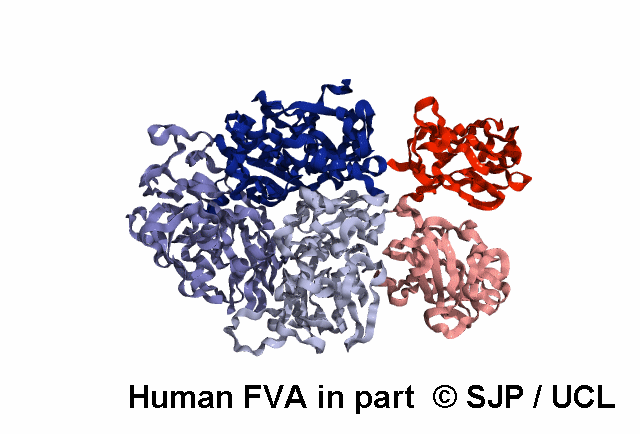
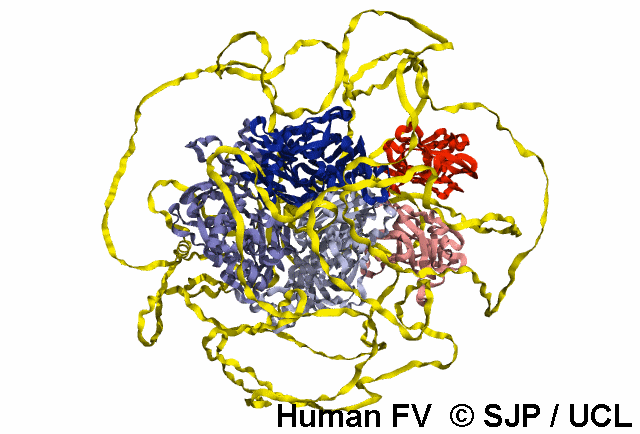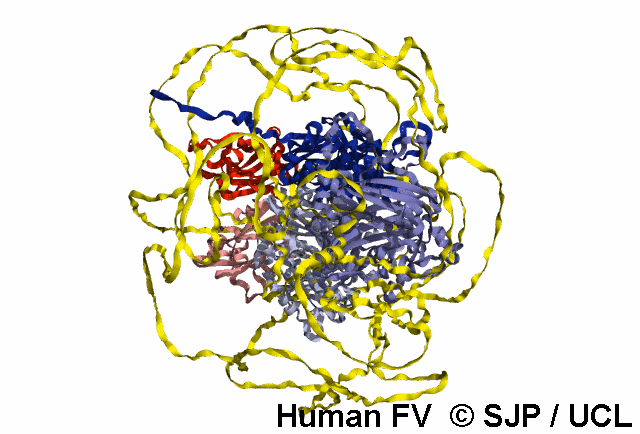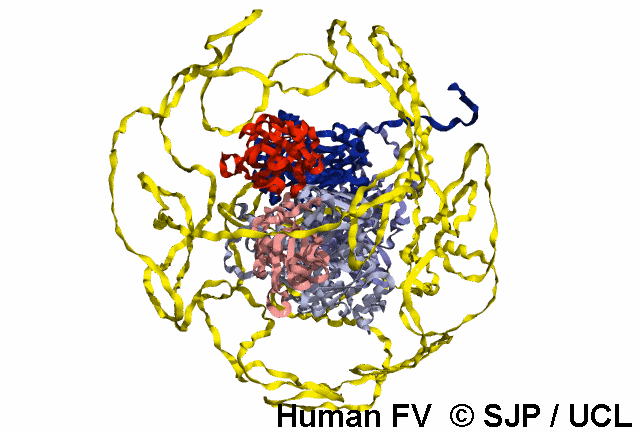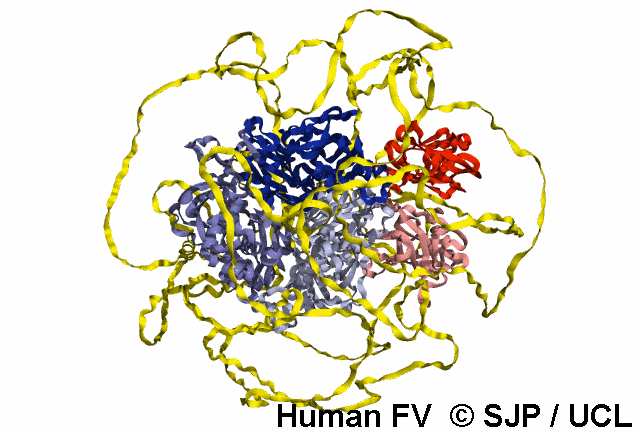
In-Depth Variant Analysis: c.43G>A (p.Gly15Ser)
Terms with a '*' next to them are explained on the Help Page .
c.43G>A
p.Gly15Ser (Legacy AA No. -13)
Variant Type:
Point
Domain:
Signal Peptide
Location:
Exon 1
Variant Effect:
Missense
Codon Change:
43G>A
No. of Patients Reported:
3
Phenotype:
NA
Allele Count *:
1735
Allele Number *:
282320
Allele Frequency *:
0.006146
Variant Comments & Reference:
NAResidue Information:
| Name | Type | Cyclic | Size | Position | Hydrophobicity | Charge | |
|---|---|---|---|---|---|---|---|
| |
|
|
|
|
|
|
|
| |
|
|
|
|
|
|
|
Substitution Analysis:
Structural Implications:
NOTE: If no Java applet appears or a Java error message is shown, please click the following LINK.